5.7 EMP_modify_assay
In general, the module EMP_identify_assay can effectively process experimental data. However, in some special cases, manual adjustment of the data is required to solve the problem.
5.7.1 Adjusting Genomic Data
In gene expression data, it sometimes happens that the expression of certain characteristics is all zero, which may cause some problems. To preserve as much information as possible, these zeros are sometimes replaced with a minimal non-zero value. This preserves the structure of the data to some extent while avoiding errors in numerical calculations.
🏷️Example1:Use the module EMP_assay_extract to replace zeros in the assay of geno_ec with a pseudo-count of 0.0001.
Before adjusting:
MAE |>
EMP_assay_extract('geno_ec')

After adjusting:
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_modify_assay('==0',pseudocount=0.0001)

🏷️Example2:Use the module EMP_modify_assay to convert absolute abundance values less than threshold 10 in the assay of geno_ec to pseudocount 5.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_modify_assay('<10',pseudocount=5)

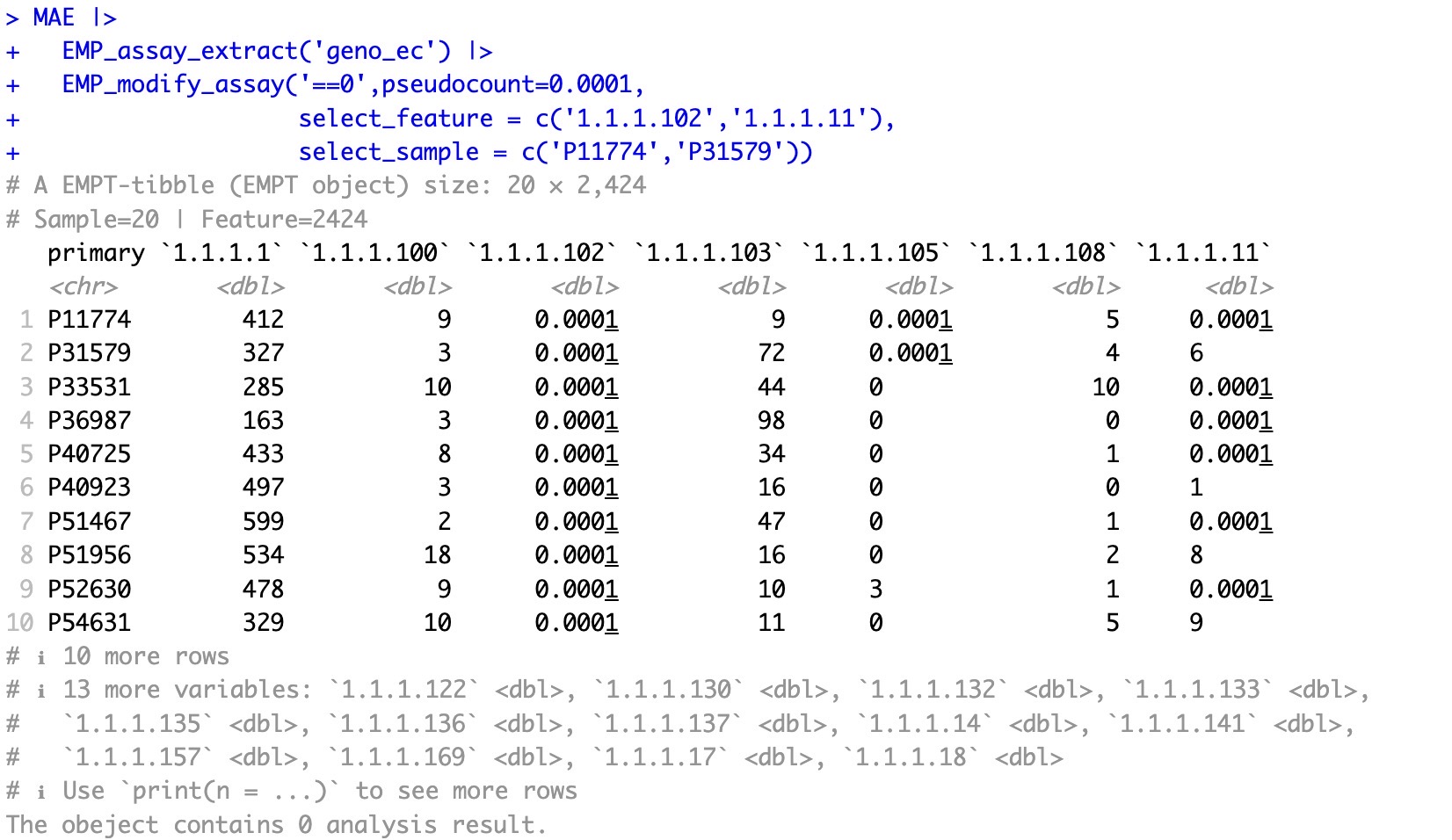
🏷️Example3:Select specific samples and features for data adjustment.
The parameters
select_feature and select_sample can be adjusted for specific sample and feature.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_modify_assay('==0',pseudocount=0.0001,
select_feature = c('1.1.1.102','1.1.1.11'),
select_sample = c('P11774','P31579'))
🏷️Example4:Suport external function.
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_modify_assay('>0',pseudocount=function(x) log(x) )

5.7.2 Adjusting Microbial Species Annotation Data
Microorganism species annotation data often contain species with minimal abundance, which may be caused by contamination, sequencing, or database alignment errors. Sometimes it is necessary to convert these extremely abundant data to zero.
This example is only for module function demonstration. To filter microbial species annotation data, please refer to 5.6 EMP_identify_assay.
🏷️Example: Use the moduleEMP_modify_assay to convert data with a relative abundance of less than 0.001 of a genus level to 0.
Before adjusting:
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_collapse(estimate_group = 'Genus',collapse_by='row') |>
EMP_decostand(method = 'relative')

After adjusting:
